Want to work with MS files yourself but do not have access to licensed software? No worries! These days, a lot of free-to-use proteomics tools (academic use mostly) are now available for mass spectrometry data analysis. However, not all programs can analyze native file extensions from different instrument vendors, such as .raw, .wiff, or .d. Luckily, the awesome proteomics community has created standardized file formats like .mzML and .mzXML, which enable cross-platform analysis. Even luckier, there is a tool that lets you convert vendor-specific extensions to these open formats!
Download now >> ProteoWizard (nope, not sponsored)
It works on all major platforms; choose it at your own will. After download, just install normally. You will also installed SeeMS software that can visualize your .mzML file.
MSConvert comes with both a GUI and a command line version, here is a list of supported format. The GUI version looks like this.
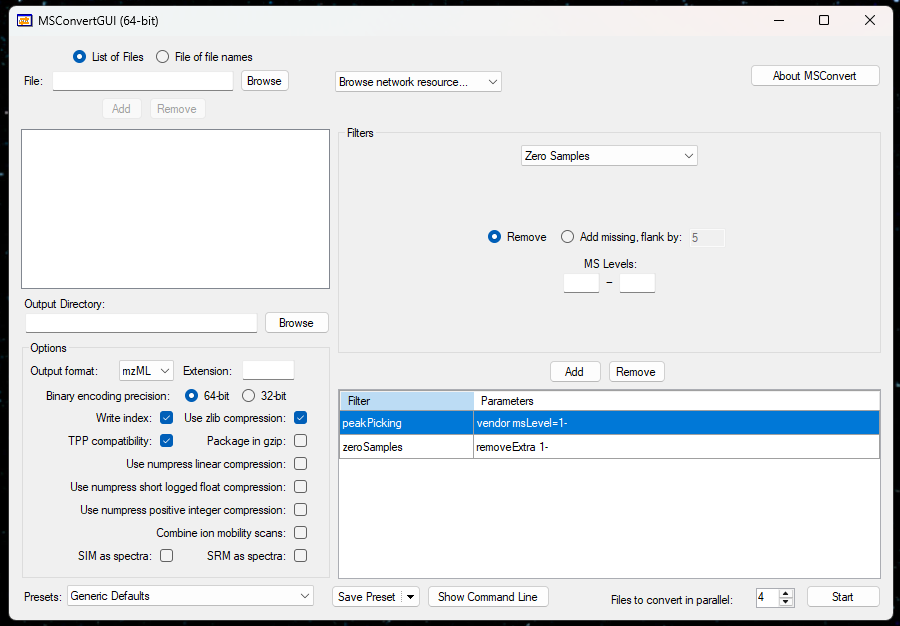
To convert files, choose your desire files and output directory. Then tick everything as in the figure and you are good to go. To add a little explanation:
- peakPicking does centroid the spectrum for you. The 1- means you do it at MS1 level.
- zeroSamples removeExtra Remove any zero data. This helps reduce file size, as .mzML normally has a larger file size than vendor-specific files.
For someone who loves the command line. You can run it by locating msconvert.exe file in the installed folder and executing it. You can also add them to PATH for more convenience. These are some of the commands I normally use, note that I use PowerShell in this setting. You can change to your platform, which should not be difficult anyway, or just ask Mr. Sam Altman to convert the format for you.
All files in the folder
-v stands for verbose. I recommend adding this argument, as it will allow you to track the progress of the conversion.
msconvert *.raw `
--filter "peakPicking true 1-" `
--mzML --64 --zlib -o .\yourfolder -v Specific files, e.g. first 10 files
In this case, we have to do it in loop.
(gci | Select-Object -first 10).BaseName |
foreach {msconvert $_ --filter "peakPicking true 1-" --mzML --64 --zlib -o .\mzML -v} Demultiplex
If by any chance you have data with staggered windows, you also need to demultiplex it.
msconvert *.raw `
--filter "peakPicking true 1-" `
--filter "zeroSamples removeExtra" `
--filter "demultiplex optimization=overlap_only" `
--mzML --64 --zlib `
-o .\yourfolder -v Happy analyzing!