For someone who works with protein, you should once in a lifetime search for the protein name to find its function. Doing so, odds are, that your search will land on the UniProt database. However, UniProt’s utility is not just to search for individual protein functions – there are tons of features embedded into this fantastic website. This section will give you a quick look at what UniProt can do beyond just naming your favorite keratin.
What Is UniProt? Swiss-Prot, TrEMBL, and UniRef Explained
Before we dive into the UniProt website itself, let’s talk about the structure of this particular database. It consists of three main components, with each serving different purposes.
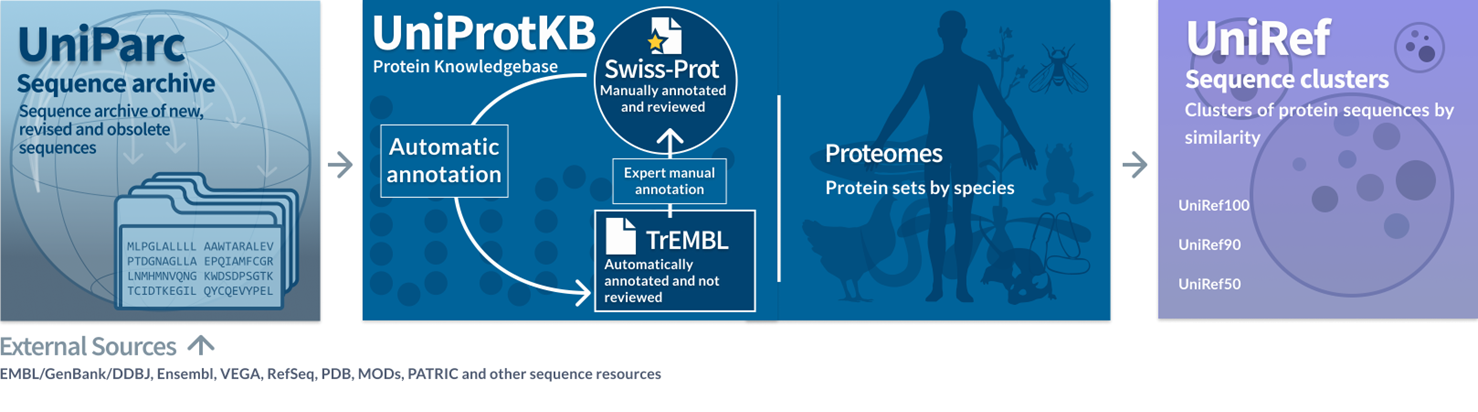
- UniParc: This is where all sequences sourced from available databases worldwide are stored. You usually won’t have to interact with this archive since it hasn’t been confirmed for its correctness yet.
- UniProtKB: This is the main part where you will interact the most. It contains two main parts: TreMBL, which contains the proteins that are computationally annotated, but no experiment data in the scientific literature yet; and Swiss-Prot, which is a manually curated database supported by evidence and literature. This is normally and subsequent process from TreMBL annotation.
- UniRef: This part contains clusters of proteins that share similar identities. If you are looking for a set of proteins with similar sequences, this is where to go.
So, unless you work on specific experiments, UniProtKB is the database you usually interact with the most.
Enough for the technical aspect, let’s move on to what this website can do!
UniProt Website
When you enter the UniProt website, you will land on its main page, which features a search bar. If you are looking for specific protein(s), you can directly search with this toolbar.
Just put the name of the protein that you want, and it will display a list of proteins that match your interest.
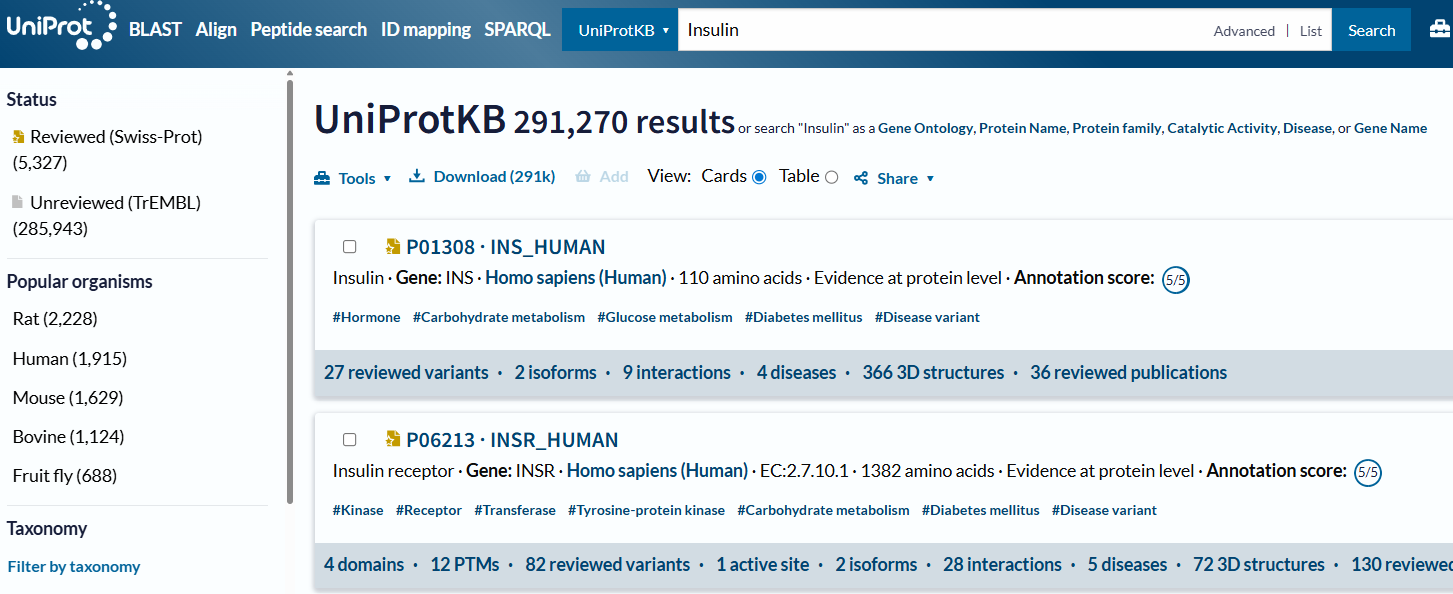
💡Tip: You can narrow down your search results by filtering with the left-hand bar panel.
That’s the most common usage of this site, but if you are wondering what else you can do, there’s the navigation bar at the top showing available tools and features.
BLAST: Used to find proteins with similar sequences. It essentially performs a BLASTP search—when you input a FASTA sequence, it searches for and returns similar protein sequences.
Align: You can input multiple sequences into the panel, and it will align them for you based on similarities and differences.
Peptide search: Similar to BLAST, but this time it finds the exact sequence (and faster).
ID mapping: Map Uniprot ID to other entries, or vice versa.
SPARQL: Programmatic assessment (SQL query). This is where the nerd is, like me.
💡Tip: For mass spectrometry folks, if you want to search your spectral data with your favorite software, you will surely need the FASTA database. Fortunately, we don’t have to manually search for all proteins within that species ourselves. There’s a panel that we can use to download the complete proteome for your chosen organism. For example, I want to search for the human proteome FASTA sequence.

Press search and look for the entries with the highest counts and relevant names. Click to open its proteome page.
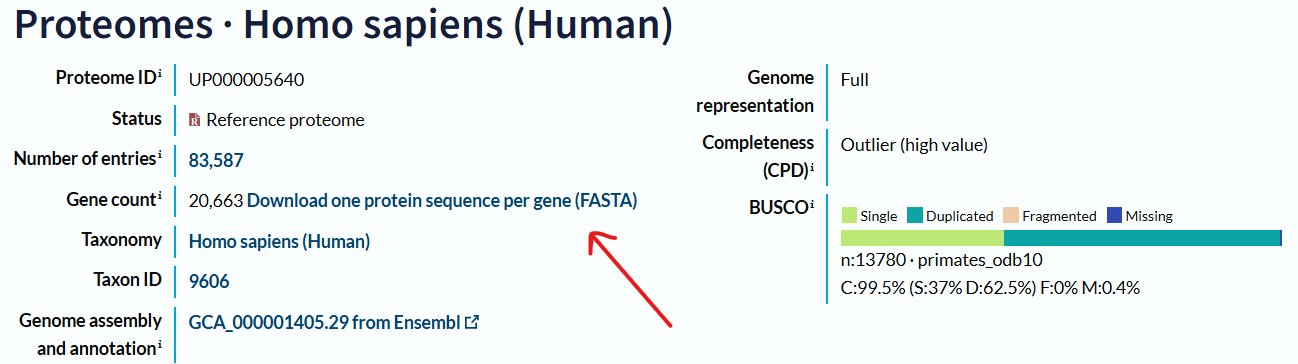 Now click “Download one protein sequence per gene (FASTA)” and get your favorite FASTA on your device!
Now click “Download one protein sequence per gene (FASTA)” and get your favorite FASTA on your device!